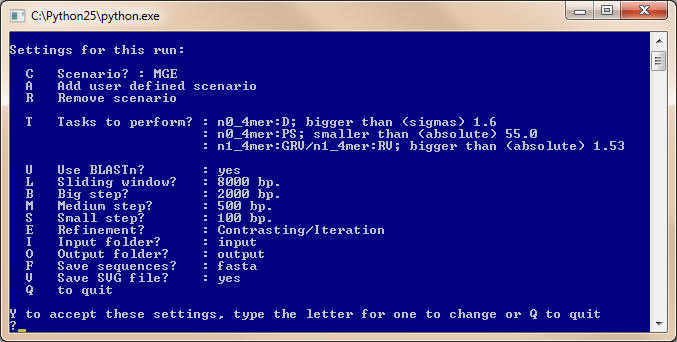
Options for the run and preset scenario
Input and output files
Options to improve prediction
Editing the task list
Setting task conditions
Addition of a new task
Save new scenario
Setting the size of the sliding window
Input and output folders
Publications

- Text output file (extension OUT);
- FASTA file of the selected genomic fragments (extension FAS), toggle by <F>;
- GenBank file for each selected genomic fragments with the annotation data (extension GBK), toggle by <F> but available only when a source GenBank file is processed;
- Graphical file with selected fragments mapped over the circular chromosome (extension SVG), toggle by <V>.
Text output file
The output files contain information about all genomic fragments enriched with the genes of interest as in the following example:
<GI> NC_013209:1 <COORDINATES>
187675-206574 <STAT> n1_4mer:GRV/n1_4mer:RV = 2.318036; n0_4mer:D =
39.633998; n0_4mer:PS = 32.759776
[187675:188712:rev]
[188776:190215:dir] DDE (Asp,Asp,Glu)
domain
[190358:191128:rev]
[191151:191543:rev]
[191715:193154:rev] DDE (Asp,Asp,Glu)
domain
[193501:194562:dir]
[194598:195656:dir]
[195653:196735:dir]
[196840:197349:rev]
[197529:199139:rev] IS66
[199203:199550:rev] IS66
Orf2
[199547:199906:rev]
[199971:201032:rev]
[201073:201156:rev] codon
recognized: UUA
[201228:201905:rev]
[201971:202426:rev]
[202423:202860:rev] ABC
transporter
[202842:203954:rev] ABC
transporter
[203990:205105:rev] ABC
transporter
[205179:205481:dir]
[205521:205946:dir]
[206080:206574:dir]
<END>
<GI> NC_013209:2 <COORDINATES>
1925341-1958404 <STAT> n1_4mer:GRV/n1_4mer:RV = 1.596704; n0_4mer:D =
37.029222; n0_4mer:PS = 21.976865
[1925341:1928319:dir]
ATP-dependent
[1928338:1928541:rev]
[1928685:1929353:rev]
[1929486:1929857:dir]
[1929911:1931551:dir]
[1931846:1934050:dir]
HAD-superfamily, FkbH domain protein
[1934292:1934651:dir]
[1934648:1934995:dir]
IS66 Orf2
[1935059:1936669:dir]
IS66
[1936858:1937937:rev] DDE
(Asp,Asp,Glu) domain
[1938092:1939906:dir]
Chromosome variant locus=SNP03 A9 no-mutation
[1940045:1940872:rev]
Integrase core domain
[1940869:1941132:rev]
[1941349:1942734:dir] DDE
(Asp,Asp,Glu) domain
[1942877:1943536:rev]
Probably pseudogene, N-terminal domain of a gene truncated by transposon
[1943835:1944566:rev]
Capsular polysaccharide biosynthesis protein
[1944581:1946164:rev]
[1946180:1946593:rev]
HAD-superfamily, Capsule biosynthesis
[1947706:1948101:dir]
[1948098:1949537:dir] DDE
(Asp,Asp,Glu) domain
[1949743:1950129:dir] DDE
(Asp,Asp,Glu) domain
[1950453:1950908:rev]
[1952377:1953762:dir] DDE
(Asp,Asp,Glu) domain
[1954741:1956429:rev]
[1956456:1957319:rev]
Polysaccharide synthesis enzyme
[1957346:1958404:rev]
<END>
In this example 2 genomic islands were identified in the genome
NC_013209.
Each block starts with the island ID, its coordinates in the genome and OU
parameters calculated for this DNA fragment. If a GenBank file was processed, the annotation and coordinates [left :
right : strand] of all genes inside the genomic fragment will be listed. The end
of the block is marked by <END>.
Graphical output file
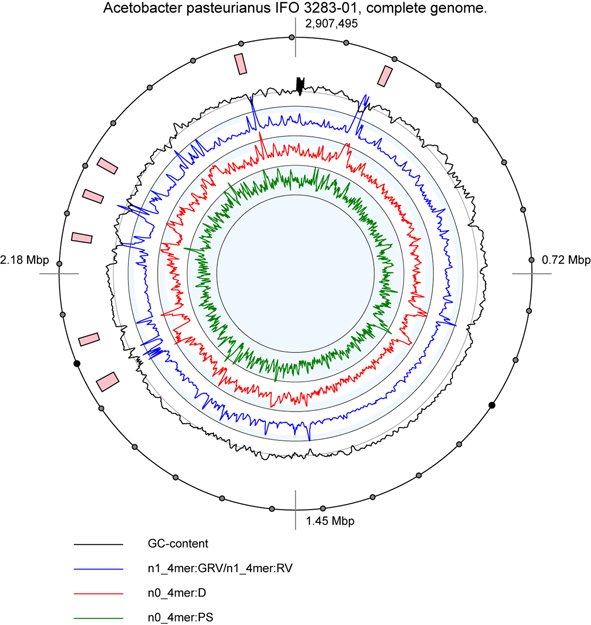
An example of the graphical output file. Pink blocks show positions of predicted genomic loci of interest.
- <U> - Use BLASTn. If set on, the program uses blastn algorithm and a small database of 16S rRNA sequences to check whether the selected genomic fragments contain rrn clusters. This option is available only for the scenarios "MGE" and "Ribosomal RNA". In the first scenario the predicted MGE is rejected if it contains rrn; in the second scenario a genomic fragment is selected only if it contains rrn.
- <E>
- Refinement. This key toggles between No |
Contrasting | Iteration | Contrasting/Itration
- Contrasting is applicable only when a GenBank file is processed. The program calculates the reference OU pattern only for the coding part of the genome and only for genes which are not annotated as hypothetical or unknown. This is a useful option when searching for mobile genetic elements;
- Iteration option instructs the program to identify genomic islands in two cycles. The loci found in the first round are excluded from the complete genome sequence and the program re-calculates the reference OU pattern.
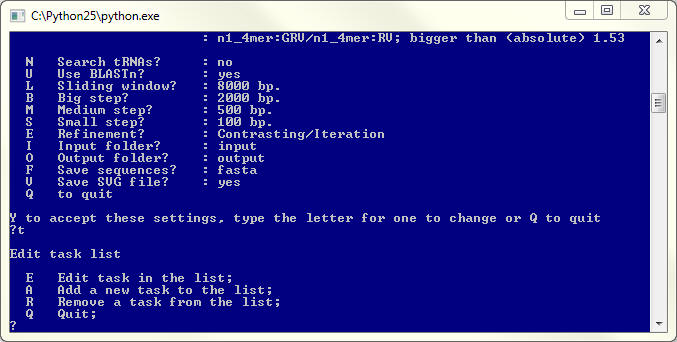
Each task is presented by a line defining the task category and the condition used to select the genomic fragments. Remember that the fragment will be selected only if it meets all set conditions. To remove a condition press <R> + <Enter>, then select the number of the task to remove it from the list.
To return to the main menu press <Q> + <Enter>.

Use the option <M> to choose the type of the threshold values:
- sigmas - to set the threshold values in sigmas of the normal distribution;
- fraction - to set the threshold as a fraction of the total number of genomic fragments;
- absolute - to use as the threshold an absolute value of the OU statistical paramenetrs.
To choose the type of comparison,- bigger than,
smaller then or between, - press the key <G>,
<S> or
<B>
respectively and press <Enter>. The program will prompt to enter the
values of one or two (if the option Between is used) thresholds. To choose
values of thresholds consult the SeqWord Browser program
(http://seqword.bi.up.ac.za//mhhapplet.php) as in the examples below:


- To choose the task category press <C>+ <Enter> and choose from the list:
- 0. return back to the previous menu;
- 1. GRV (generalized relative variance);
- 2. PS (pattern skew);
- 3. RV (relative variance);
- 4. D (pattern deviation - by default);
- 5. GCS (GC-skew);
- 6. GD (generalized pattern deviation);
- 7. GC (GC-content);
- 8. AT (AT-content);
- 9. GPS (generalized pattern skew);
- 10. ATS (AT-skew); (for more about OU statistical parameters see Reva and Tümmler, 2005)
- To change the oligonucleotide word length press <W>+ <Enter> and enter an integer from 2 to 7 (4 by default).
- To set the normalization press <N>+<Enter> and enter an integer from 0 (no normalization) to word_length - 1. (Normalization by the mononucleotide content of the sequence, - option 1, - is set by default. Remember, that when generalized parameters are selected, - GRV, GD or GPS, - for normalization the frequencies of the complete genome are taken into consideration, whereas by default the parameters are normalized by the content of the genomic region selected by a sliding window.)
- The program allows execution of simple mathematical operations with the OU statistical parameters such as subtraction and division (or [par1-par2]/par3 if the subtrahend (par2) and the divisor (par3) are both set). Thus, in the scenario of identification of horizontally transferred gene islands the program calculates deviation n1_4mer:GRV/n1_4mer:RV - this ratio is around 1.0 for the core sequence but higher than 2 in genomic fragments from the accessory genome. (When setting the divisor be sure that this parameter is never zero!) To set subtraction or division of the parameters, press correspondingly <S>+<Enter> or <D>+<Enter>. The program will show a menu similar to the discussed above menu for addition of a new task.
Press <A>+<Enter> to add a subtrahend
or a divisor, or to add the new task to the list. In the letter case the
program will show the condition setting menu that was described above.
Press <Q>+<Enter> to return to the task edit menu and again
<Q>+<Enter> to return to the main menu.
To change the values of the sliding window length (8 000 bp), big step (2 000 bp), medium step (500 bp) and small step (100 bp) set by default, press the keys <L>, <B>, <M> and <S> correspondingly and press <Enter>. The program will prompt you to enter new values. (Remember that for statistical reliability the sliding window size should not be shorter than 4600 bp for tetranucleotide usage analysis, 1200 bp for trinucleotides and 600 bp for dinucleotides.
- Ganesan H, Rakitianskaia AS, Davenport CF, Tümmler B, Reva ON. (2008) The SeqWord Genome Browser: an online tool for the identification and visualization of atypical regions of bacterial genomes through oligonucleotide usage. BMC Bioinformatics. 9:333.
- Reva, O.N., Tümmler, B. (2005). Differentiation of regions with atypical oligonucleotide composition in bacterial genomes. BMC Bioinformatics. 6:251.
- Reva O., Tümmler B. (2008) Think big - giant genes in bacteria. Environ. Microbiol. 10(3), 768-777.