Back to SeqWord Project main page
Back to Barcoder Help file
Download examples of artificial metagenomes
Download program Barcoding 2.0
Barcoding 2.0
The program Barcoding 2.0 is a command line program on Python 2.5 designed to align metagenome reads
(Roche 454 and Illumina) against taxon specific barcode sequences generated by the on-line program BarcodeGenerator.
To download the program, click here [320 MB].
To download examples of generated taxon specific barcode sequences, click here.
To download examples of artificial metagenomes, click here.
Download ZIP file of Barcoding 2.0 to your computer and unzip it. You will see the following folders:
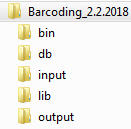
Do the following steps:
- Copy following files to the folder 'input':
- FASTA file with barcode sequences. Let it be named 'barcodes.fas'. Several examples of generated barcodes are available here
- FASTA file of metagenomic reads. If you have several metagenomes, create an new folder 'metagenomes' in the folder 'input' and copy metagenome files there.
Several examples of generated artificial metagenomes are available here
- Optionally, copy to the folder 'input' a phylogenetic tree in Phylip/Newick format to align barcoded taxa against the phylogenetic tree. Taxonomic units in the tree file MUST have
the same names as in the barcode file. At least 3 barcoded taxa should be present in the tree file.
Hint: you may generate a phylogenetic tree directly from the initial GenBank file by means of our program SWPhylo
- Run the program by double-clicking on the file 'run.py' in the top level folder of the program. Command line interface of the program is shown below:
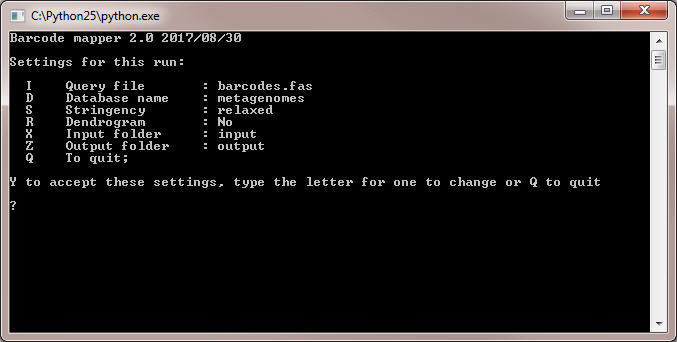
- Check that the names of Query file (barcode file) and Database name (name of the subfolder containing FASTA files with metagenomic reads)
correspond to the real names in the folder 'input'. Otherwise change the names using keys I and D, respectively.
- Use the option S to choose Stringency parameter between 'relaxed' and 'stringent'.
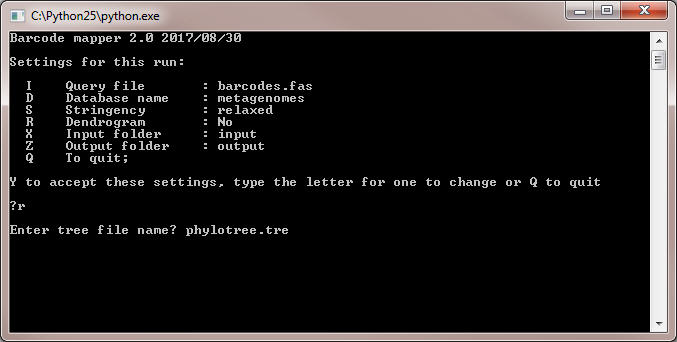
- If you have to use a phylogenetic tree file, let's say it is named 'phylotree.tre', copy it first to the folder 'input'.
- Type F and press Enter. The program will ask you to enter the name of the tree file:
- If program does not find the indicated file in the folder 'input', no dendrogram option will be returned. If the file does exist, its name will apear in the option set:
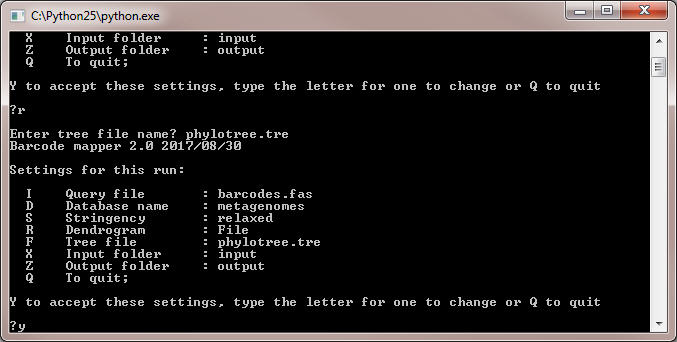
- Type Y end press Enter. The program will start showing the progress.
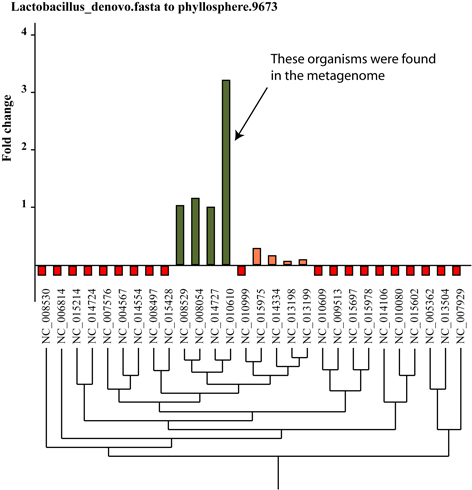
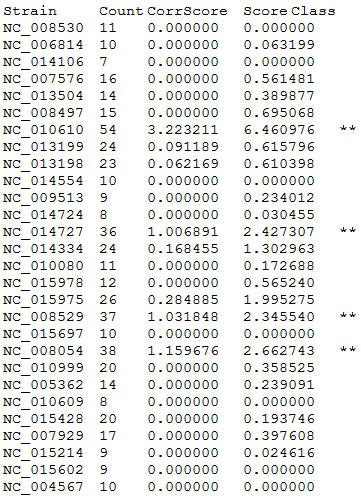
- The program will generate two files shown beow:
- Graphical file in SVG format;
- Text file;
Contact Information
Company Name: University of Pretoria
Website: http://bargene.bi.up.ac.za/
Email: oleg.reva@up.ac.za;
Phone number:+27-12-420-5810






