Back to SeqWord Project main page
Back to Barcoder Help files
Download diagnostic barcodes
Download an input example (zipped file of 8 Bacillus genomes - 21 kb)
BarcodeGenerator
Download BarcodeGenerator (Python 2.5 or 2.7) [48 kb]
How to use BarcodeGenerator On-Line
The program BarcodeGenerator allows for the creation of barcode sequences based on a given set of genomes, it compares all pairs of genomes and select barcodes (DNA sequences) out of the core genes.
The program allows the addition of accessory genes which are genome specific to improve the sensitivity of the barcode sequences.
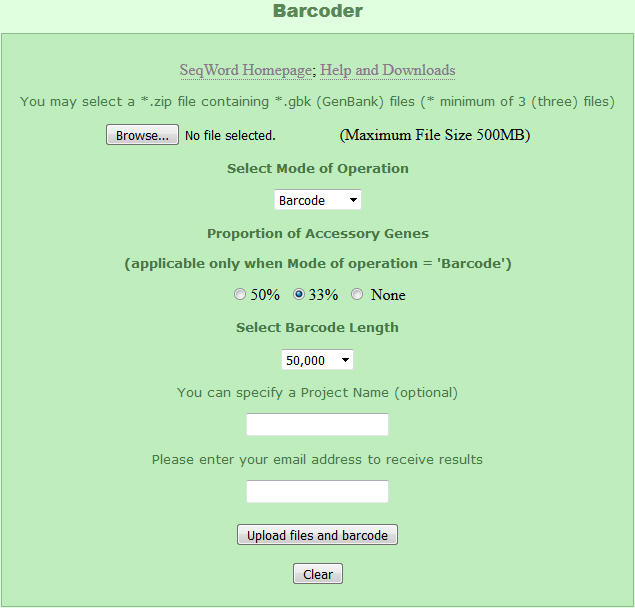
Perform the following steps:
- Click on "Browse" and choose a zipped file with a collection of genomes of interest in GenBank format;
- For barcode generation, leave 'Barcode' as a mode of operation. Other options are discussed below;
- Select proportion of accessory genes in generated barcode sequences;
- Select an approximate length of barcode sequences;
- Enter the prject name;
- Enter your e-maile address to receive a link to the file with generated barcode sequences;
- Click on "Upload files and barcodes" to start the task;
- Check your mail box for a message on the process completion;
How to use the local version of BarcodeGenerator
The program BarcodeGenerator is a command line program on Python 2.5 designed
to select the most appropriate genes for genetic barcoding and generate barcode sequences, which then can be used for analysis and visualisation of
metagenomic datasets by using the program Barcoding 2.0.
To download the program, click here [48 kb].
To download examples of generated taxon specific barcode sequences, click here.
To download examples of artificial metagenomes, click here.
Download ZIP file of BarcodeGenerator to your computer and unzip it. You will see the following folders:
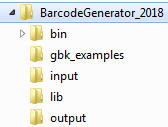
Do the following steps:
- Copy GenBank files of organisms you want to barcode to the folder 'input':
- Run the program by double-clicking on the file 'run.py' in the top level folder of the program. Command line interface of the program is shown below:
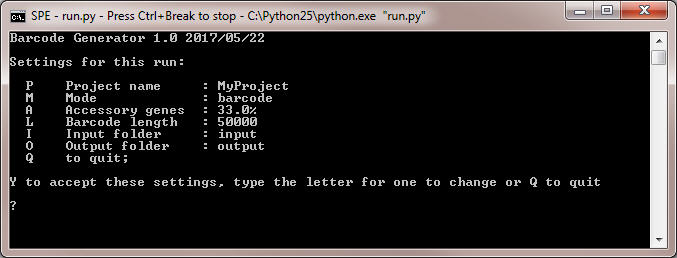
- Use option P to change the project name.
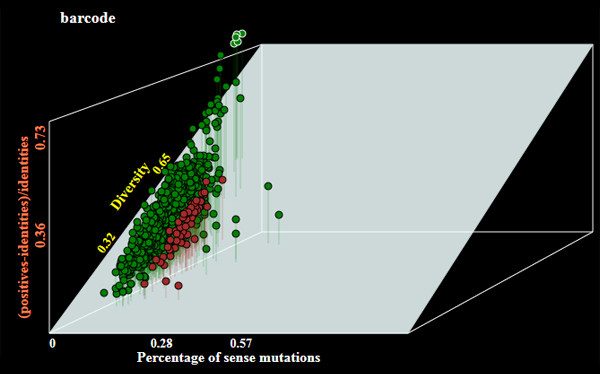
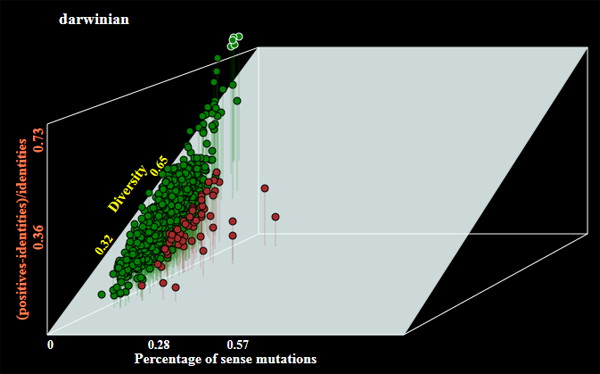
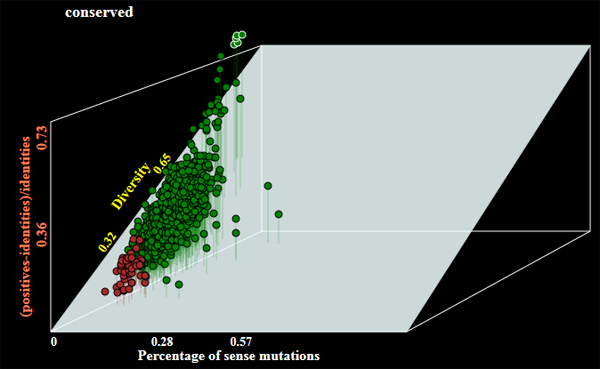
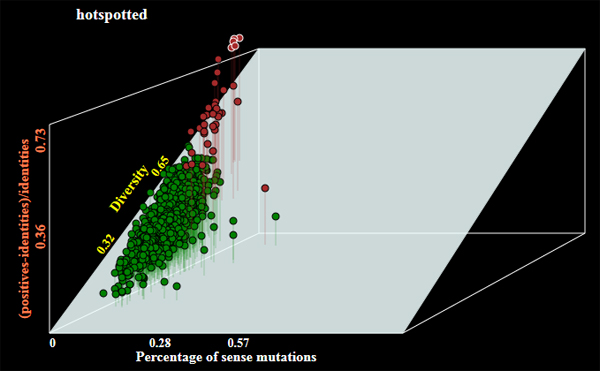
- Operation mode by default is barcode to generate barcode sequences. The program may allow selection of genes of the following alternative categories:
- darwinian - orthologous genes under positive selection;
- conserved - the most conserved genes;
- hotspotted - orthologous genes with highest number of random mutations;
Examples of genes selected under different modes of operations are shown below:
- Use option A to set the percentage of accessory genes in barcode sequences. This option is available only with the barcode mode of operation;
- Use option L to set an average length of barcode sequences in bp;
- Type Y end press Enter. The program will start showing the progress.
- The program will generate 3 output files and store them in the folder 'output':
- FASTA file of barcode sequences;
- Graphical output with a plot of selected genes (see examles above);
- Text report;
Examples of generated barcodes see here
Contact Information
Company Name: University of Pretoria
Website: http://bargene.bi.up.ac.za/
Email: oleg.reva@up.ac.za;
Phone number:+27-12-420-5810