Assigning sequences to functional categories
(!!!NEW For automatic identification of genomic regions belonging to different functional categories use SeqWord Sniffer!)
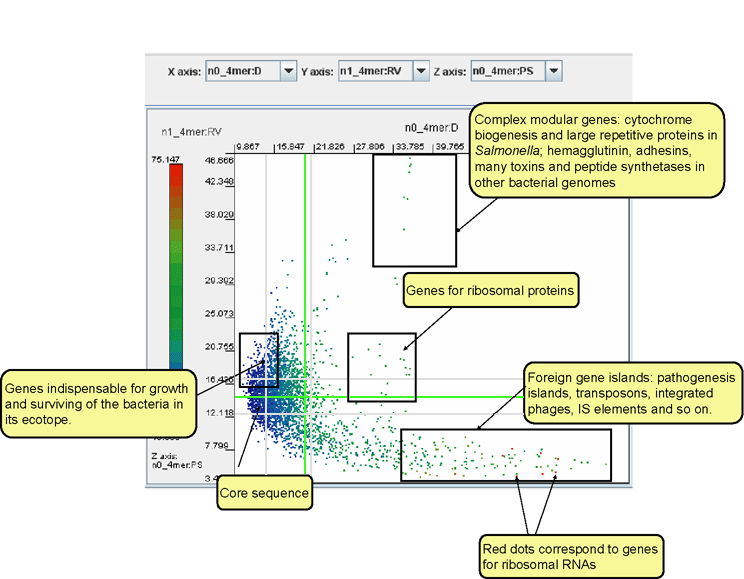
Genomic loci can be putatively assigned to different functional categories based upon the tetranucleotide parameters distance, variance RV and pattern skew PS. Functional categories include so called fitness genes (genomic fragments predicted to be highly selected for in the organism), foreign genomic islands, ribosomal proteins, or the core genome.
To analyze local variations of OU patterns in bacterial genomes and identify genomic loci of different functional categories:
Go to the tab ‘Diagram’
Select n0_4mer:D for the X axis, n1_4mer:RV for the Y axis and n0_4mer:PS for the Z axis.
Click ‘Enter’.
To better navigate around the dot plot, display inner quartiles and mean values for the axes X and Y by selecting the correspondent checkboxes on the panel ‘Statistical data’. The distribution of genomic fragments of different functional categories is shown on the image below.

Bulk genomic fragments are distributed left of the vertical mean-value line.
The clusters of genes for ribosomal proteins are presented as a cloud of dots above the horizontal mean-value line and to the right from the second (right) vertical inner-quartile line.
The upper part of the window (from the second (upper) horizontal and second (right) vertical inner-quartile lines) contains long modular genes (polypeptide synthases, surface adhesion proteins, etc). Non-coding sequences with multiple tandem repeats are in the same area but above and to the right of multidomain genes.
The lower-right sector below the first (lower)
horizontal and second
(right) vertical inner-quartile lines comprises the genomic islands and
pseudogenes - non-coding
remnants of former genes. The bright red dots in the same sector depict
the
clusters of genes for ribosomal RNAs.
Due to their length and frequent repeats in these genes, tetranucleotide patterns are distinct from the genomic average. Also, with RV being high there is selection for their conservation and against random mutations. This technique is explained in full with examples in a previous publication (Reva and Tümmler 2008).